How To Structure Data Science Projects
Structuring a data science project is very important to logically conduct experiments, effectively share your findings with colleagues and easily reproduce your work. It saves a lot of time by removing any confusion and improving information sharing. In this blog post, I summarize a strategy to structure a data science project and some tips to organize codes. Let’s get started!
Structuring a data science project involves has 3 parts:
- Folder management
- Data management
- Code management
1. Create an appropriate folder structure
- Make a separate folder for each project
- Keep logical folder names
- Each folder should contain files of only one type
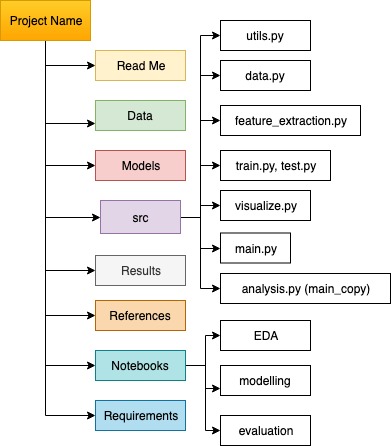
Example folder structure
1. LICENSE
2. README.md
3. requirements.txt
4. dataset
4.1 raw
4.1.1 train
4.1.2 test
4.2 processed
4.3 interim
4.4 external -> data from external sources
5. models -> save the trained models
summary.txt -> summary of all models (parameters, conditions) in a text file
6. results -> save the results by date ex. YYYYMMDD.model_name.prediction.jpg
7. src
7.1 __init__.py -> Makes src a Python module
7.2 data
make_dataset.py
7.3 features
build_features.py
7.4 models
train.py
predict.py
7.5 visualization -> EDA, model and result visualization
visualization.py
8. notebooks -> for exploratory data analysis and explain the flow to others
9. references -> papers, online blogs, articles etc.
10. set_up.py -> to make the project pip installable
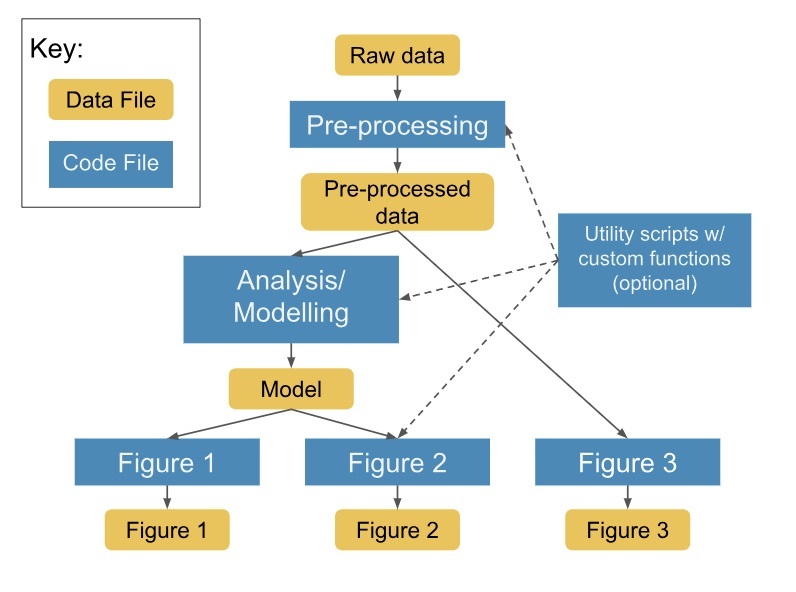
2. Sketch the flow of data and code
- Create a diagram that shows the interaction of data and code before development
- Discuss your code structure with a friend to check if it is easily understandable
Example diagram (credits: Kaggle)
3. Modularize the code as much as possible
- Make three types of scripts :
- general function scripts,
- custom function script for each type of functionality,
- main script to run the entire code
- Break the code into smaller pieces each intended to perform a specific task. If the code has 5 main blocks, divide the code into 5 python files. Then in the main script, call functions from each file sequentially.
- Each file should have functions of same functionality/type/use - data, model, visualization etc.
- Group the functions into modules (or python files) based on its usability. It also helps in staying organized and ease of code maintainability
4. Make your code readable
- Write your code like you write English.
- Put all your imports, import x or library(x) at the top of your notebook.
- Make sure to use white space and use it consistently. White space can serve like the breaks between paragraphs to help you group similar concepts together and help your readers follow the logical flow of the text.
- Break up long lines at logical places. For example, if you have a function that takes a lot of arguments, you can try inserting a line break after each argument.
- Make your variable names logical and human readable. The variable name should clearly indicate what it contains.
- Comment your code! Especially for research code, when your intended audience may not be as fluent in your language of choice.
- Use consistent patterns of capitalization. The prevailing pattern in data science tends to be ‘lower_case_with_underscores’ for functions and variables, and ‘firstlowerThencapitalize’ for classes.
- Use the doc_string format when writing functions
- Use relative rather than absolute file paths
- Avoid the following when naming files- spaces, punctuation, accented characters, file names that don’t tell anything about the content
- Refer style guide PEP 8, can use linter for formatting code
Don’t waste too much time in beautifying your code, main point is that it should be smoothly readable and easily reproducible.
5. Maintaining Research Code
- For research projects, create separate files for main code and analysis code
- Try all the tests/functions on analysis code and after analysis append the useful ones in main code
- Use Jupyter Notebooks for initial prototyping, later convert the notebook into python scripts
6. Save the environment information
-
Save the environment information so that others can easily reproduce your work in their machine and you can also port to a different setting
import IPython # print system information (but not packages) print(IPython.sys_info()) # get module information !pip freeze > frozen-requirements.txt # append system information to file with open("frozen-requirements.txt", "a") as file: file.write(IPython.sys_info())
8. Controlling randomnness
- Machine learning experiments are stochastic and have many sources of randomness.
-
Set all the random number generators your code depends upon (below slide is from Kaggle)
# Set a seed value: seed_value= 12321 # 1. Set `PYTHONHASHSEED` environment variable at a fixed value: import os os.environ['PYTHONHASHSEED']=str(seed_value) # 2. Set `python` built-in pseudo-random generator at a fixed value: import random random.seed(seed_value) # 3. Set `numpy` pseudo-random generator at a fixed value: import numpy as np np.random.seed(seed_value) # 4. Set `tensorflow` pseudo-random generator at a fixed value: import tensorflow as tf tf.set_random_seed(seed_value) # 5. For layers that introduce randomness like dropout, make sure to set seed values: model.add(Dropout(0.25, seed=seed_value)) #6 Configure a new global `tensorflow` session: from keras import backend as K session_conf = tf.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1) sess = tf.Session(graph=tf.get_default_graph(), config=session_conf) K.set_session(sess)
7. Make an easy to understand ReadMe
It should contain 5 parts:
- project objective
- example of input and output
- description of dataset
- dependencies and system information
- example to run the code
- how to cite
8. Use version control and Git (need to learn further)
9. Clean your project structure regularly
References
- http://www.rctatman.com/files/Tatman_2018_ReproducibleML.pdf
- Reproducible research best practices
- Cookiecutter data science
- How to write production level code in data science
- Example directory structure
- Properly setting the random seed in machine learning experiments
- How to manage machine learning projects
- Best Practices for Scientific Computing